checkCytoscapeVersion
Checks cytoscape version.
CheckResponse(r)
hostcytoscape host address, default=cytoscape_hostportcytoscape port, defaul=cytoscape_portreturnsnothing
>>> import AGEpy as age
>>> age.checkCytoscapeVersion()
cytoscapeVersion 3.6.0
apiVersion v1
cytoscape
General function for interacting with Cytoscape API.
cytoscape(namespace,command="",PARAMS={},host=cytoscape_host,port=cytoscape_port,method="POST",verbose=False)
namespacenamespace where the request should be executed. eg. "string"commnandcommand to execute. eg. "protein query"PARAMsa dictionary with the parameters. Check your swagger normaly running on 'http://localhost:1234/v1/swaggerUI/swagger-ui/index.html?url=http://localhost:1234/v1/commands/swagger.json'hostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portmethodtype of http call, ie. "POST" or "GET" or "HELP".verboseprint more informationreturnsFor "POST" the data in the content's response. For "GET" None.
>>> import AGEpy as age
>>> response=age.cytoscape("string","pubmed query",{"pubmed":"p53 p21","limit":"50"})
>>> print response
{u'SUID': 37560}
result
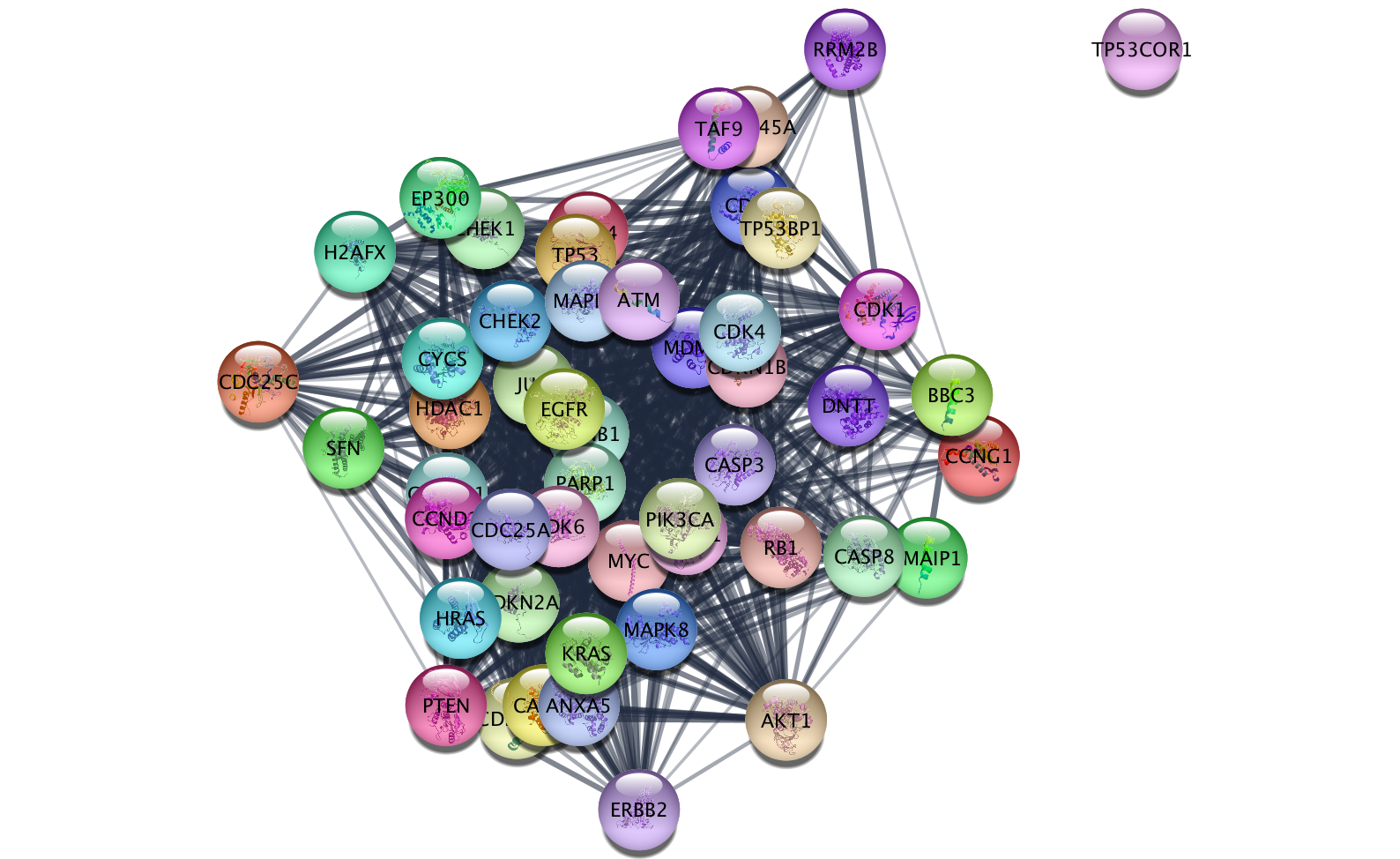
Displays the current network.
result(filetype="PNG", saveas=None, host=cytoscape_host, port=cytoscape_port)
filetypefile type, default="PNG"saveas/path/to/non/tmp/file.prefixhostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portreturnsan image
>>> import AGEpy as age
>>> response=age.result()
>>> response
getTableColumns
Gets tables from cytoscape.
getTableColumns(table, columns, namespace = "default", network = "current", host=cytoscape_host,port=cytoscape_port,verbose=False)
tabletable to retrieve eg. nodecolumnscolumns to retrieve in list formatnamespacenamepsace, default="default"networka network name or id, default="current"hostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portverboseprint more informationreturnsa pandas dataframe
>>> import AGEpy as age
>>> response=age.getTableColumns('node',['display name'])
>>> print response
display name
9606.ENSP00000367207 MYC
9606.ENSP00000356150 MDM4
9606.ENSP00000228872 CDKN1B
9606.ENSP00000361021 PTEN
9606.ENSP00000265734 CDK6
loadTableData
Loads tables into cytoscape.
loadTableData(df, df_key='index',table="node", table_key_column = "name", network="current", namespace="default", host=cytoscape_host, port=cytoscape_port, verbose=False)
dfa pandas dataframe to loaddf_keykey column in df, defaul="index"tabletarget table, default="node"table_key_columntable key column, default="name"networka network name or id, default="current"hostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portverboseprint more informationreturnsoutput of put request
>>> import AGEpy as age
>>> print df.head()
display name
9606.ENSP00000367207 MYC
9606.ENSP00000356150 MDM4
9606.ENSP00000228872 CDKN1B
9606.ENSP00000361021 PTEN
9606.ENSP00000265734 CDK6
>>> def MarkCKDs(x):
... if "CDK" in x:
... res="yes"
... else:
... res="not"
... return res
>>> df["CDK"]=df["display name"].apply( lambda x: MarkCKDs(x) )
>>> print df.head()
display name CDK
9606.ENSP00000367207 MYC not
9606.ENSP00000356150 MDM4 not
9606.ENSP00000228872 CDKN1B yes
9606.ENSP00000361021 PTEN not
9606.ENSP00000265734 CDK6 yes
>>> response=age.loadTableData(df[["CDK"]])
simple_defaults
Simplifies default layouts.
simple_defaults(defaults_dic)
defaults_dica dictionary of the form { visualProperty_A:value_A, visualProperty_B:value_B, ..}returnsa list of dictionaries with each item corresponding to a given key in defaults_dic
>>> import AGEpy as age
>>> defaults_dic={"NODE_SHAPE":"ellipse",\
"NODE_SIZE":60,\
"NODE_FILL_COLOR":"#AAAAAA",\
"EDGE_TRANSPARENCY":120}
>>> defaults_list=age.simple_defaults(defaults_dic)
>>> print defaults_list
[{'visualProperty': 'NODE_SIZE', 'value': 60}, \
{'visualProperty': 'NODE_FILL_COLOR', 'value': '#AAAAAA'}, \
{'visualProperty': 'NODE_SHAPE', 'value': 'ellipse'}, \
{'visualProperty': 'EDGE_TRANSPARENCY', 'value': 120}]
create_styles
Creates a new visual style.
create_styles(title,defaults=None,mappings=None,host=cytoscape_host,port=cytoscape_port)
titletitle of the visual styledefaultsa list of dictionaries for each visualPropertymappingsa list of dictionaries for each visualPropertyhostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portretunrsnothing
>>> import AGEpy as age
>>> print defaults_list
[{'visualProperty': 'NODE_SIZE', 'value': 60}, \
{'visualProperty': 'NODE_FILL_COLOR', 'value': '#AAAAAA'}, \
{'visualProperty': 'NODE_SHAPE', 'value': 'ellipse'}, \
{'visualProperty': 'EDGE_TRANSPARENCY', 'value': 120}]
>>> response=age.create_styles("newStyle",defaults=defaults_list)
update_style
Updates a visual style.
update_style(title, defaults=None, mappings=None, host=cytoscape_host, port=cytoscape_port, verbose=False)
titletitle of the visual styledefaultsa list of dictionaries for each visualPropertymappingsa list of dictionaries for each visualPropertyhostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portretunrsnothing
>>> import AGEpy as age
>>> print new_defaults_list
[{'visualProperty': 'NODE_SIZE', 'value': 80}, \
{'visualProperty': 'NODE_FILL_COLOR', 'value': '#AAAAAA'}, \
{'visualProperty': 'NODE_SHAPE', 'value': 'ellipse'}, \
{'visualProperty': 'EDGE_TRANSPARENCY', 'value': 120}]
>>> response=age.update_style("newStyle",defaults=defaults_list)
mapVisualProperty
Generates a dictionary for a given visual property
mapVisualProperty(visualProperty, mappingType, mappingColumn, lower=None,center=None,upper=None, discrete=None, network="current",table="node", namespace="default", host=cytoscape_host, port=cytoscape_port, verbose=False)
visualPropertyvisualPropertymappingTypemappingTypemappingColumnmappingColumnlowerfor "continuous" mappings a list of the form [value,rgb_string]centerfor "continuous" mappings a list of the form [value,rgb_string]upperfor "continuous" mappings a list of the form [value,rgb_string]discretefor discrete mappings, a list of lists of the form [ list_of_keys, list_of_values ]networka network name or id, default="current"hostcytoscape host address, default=cytoscape_hostportcytoscape port, default=cytoscape_portretunrsa dictionary for the respective visual property
>>> import AGEpy as age
>>> import matplotlib
>>> NODE_LABEL=age.mapVisualProperty("NODE_LABEL","passthrough","display name")
>>> print NODE_LABEL
{'mappingType': 'passthrough', 'visualProperty': 'NODE_LABEL', 'mappingColumnType': u'String', 'mappingColumn': 'display name'}
>>> NODE_SHAPE=age.mapVisualProperty('NODE_SHAPE','discrete','CDK',\
discrete=[ ["yes","not"], \
["DIAMOND", "ellipse"] ])
>>> NODE_SIZE=age.mapVisualProperty('NODE_SIZE','discrete','CDK',\
discrete=[ ["yes","not"],\
["100.0","60.0"] ])
# imagine you have a log2(fold_change) column in your cytoscape table
>>> cmap = matplotlib.cm.get_cmap("bwr")
>>> norm = matplotlib.colors.Normalize(vmin=-4, vmax=4)
>>> min_color=matplotlib.colors.rgb2hex(cmap(norm(-4)))
>>> center_color=matplotlib.colors.rgb2hex(cmap(norm(0)))
>>> max_color=matplotlib.colors.rgb2hex(cmap(norm(4)))
>>> NODE_FILL_COLOR=age.mapVisualProperty('NODE_FILL_COLOR','continuous','log2(fold_change)',\
lower=[-4,min_color],center=[0.0,center_color],upper=[4,max_color])
aDiffCytoscape
Plots tables from aDiff/cuffdiff into cytoscape using String protein queries. Uses top changed genes as well as first neighbours and difusion fo generate subnetworks.
aDiffCytoscape(df, aging_genes, target, species="caenorhabditis elegans", limit=None, cutoff=0.4, taxon=None, cytoscape_host=cytoscape_host, cytoscape_port=cytoscape_port)
dfdf as outputed by aDiff for differential gene expressionaging_genesENS gene ids to be labeled with a diagonaltargettarget destination for saving files without prefix. eg. "/beegfs/group_bit/home/JBoucas/test/N2_vs_daf2"speciesspecies for string app query. eg. "caenorhabditis elegans", "drosophila melanogaster", "mus musculus", "homo sapiens"limitlimit for string app query. Number of extra genes to recover. If None, limit=N(query_genes)*.25cuttoffconfidence cuttoff for sting app query. Default=0.4taxontaxon id for string app query. For the species shown above, taxon id will be automatically identifiedcytoscape_hosthost address for cytoscape, default=cytoscape_hostcytoscape_portcytoscape port, defaut=cytoscape_portreturnsnothing
>>> import AGEpy as age
>>> print genes[:10]
['WBGene00008288', 'WBGene00002169', 'WBGene00008733', 'WBGene00004178', 'WBGene00004178', 'WBGene00004179', 'WBGene00004179', 'WBGene00020581', 'WBGene00001877', 'WBGene00001881']
>>> print df.head()
ensembl_gene_id gene locus sample_1 sample_2 status \
0 WBGene00022275 Y74C9A.1 I:43732-44677 N2 daf2 OK
1 WBGene00004418 F53G12.9,rpl-7 I:111037-113672 N2 daf2 OK
2 WBGene00018774 F53G12.9,rpl-7 I:111037-113672 N2 daf2 OK
3 WBGene00018772 F53G12.4 I:134336-137282 N2 daf2 OK
4 WBGene00018958 F56C11.6 I:171339-175991 N2 daf2 OK
value_1 value_2 log2(fold_change) test_stat p_value q_value \
0 0.195901 0.986634 2.332390 2.32959 0.00570 0.031216
1 3354.820000 2463.480000 -0.445539 -2.71381 0.00005 0.000556
2 3354.820000 2463.480000 -0.445539 -2.71381 0.00005 0.000556
3 1.235670 2.992460 1.276040 3.16508 0.00005 0.000556
4 2.651180 3.795600 0.517696 1.73994 0.00410 0.024157
significant GO_id \
0 yes NaN
1 yes GO:0003735; GO:0000463; GO:0044822; GO:0002181...
2 yes NaN
3 yes NaN
4 yes GO:0016787; GO:0005615; GO:0004104
GO_term gene_biotype \
0 NaN protein_coding
1 structural constituent of ribosome; maturation... protein_coding
2 NaN protein_coding
3 NaN protein_coding
4 hydrolase activity; extracellular space; choli... protein_coding
NormInt evidence
0 -0.356904 no
1 3.458609 no
2 3.458609 no
3 0.283965 no
4 0.501360 no
>>> age.aDiffCytoscape(df,genes,"/u/home/JBoucas/cytoscape/cyto")